CRISPR gene editing holds huge promise for tackling a wide range of inherited diseases, but concerns about unintended “off target” effects has slowed its rollout. A redesigned CRISPR system could finally have a practical solution to this problem.
Last month, biomedical company Intellia reported promising results from a phase one trial of a CRISPR therapy that treats a rare disease caused by malformed proteins in the liver. But almost a decade after the discovery of the gene-editing tool, there are still only a handful of therapies in clinical trials, and none have been approved by regulators so far.
One major reason for that is concern around the technology’s tendency to make significant numbers of unintentional changes to the genome that could lead to serious side effects, including cancer. There have been plenty of efforts to redesign the tool to make it more accurate, but typically this has resulted in a substantial decrease in the speed of editing.
By carefully studying the structure of the CRISPR system, a team of researchers from the University of Texas at Austin have now found a way to redesign the critical Cas9 protein in a way that reduces errors while maintaining performance. “This really could be a game changer in terms of a wider application of the CRISPR Cas systems in gene editing,” co-senior author Kenneth Johnson said in a press release.
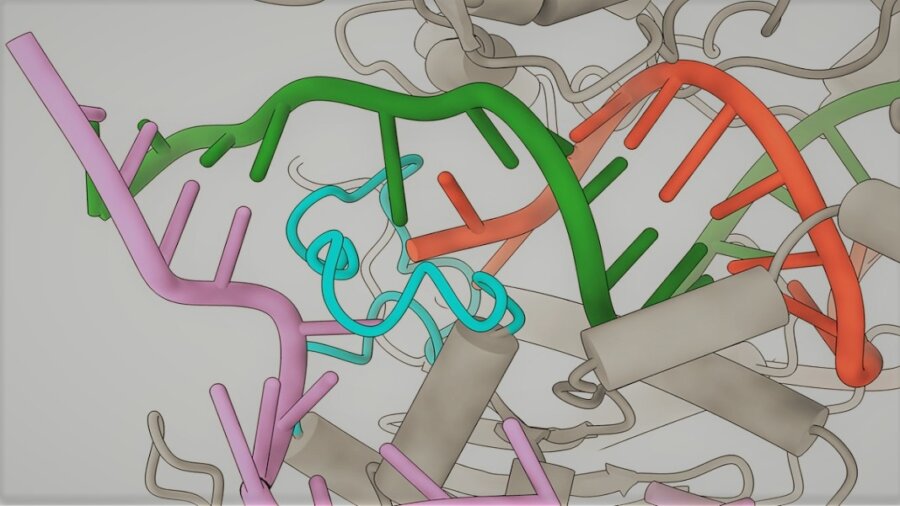
The Cas9 protein plays a central role in the CRISPR system. The enzyme is responsible for making cuts in sequences of DNA, which makes it possible to remove errant genes or make space to add new genetic material. To make sure the enzyme makes cuts in the right place, it’s paired with a piece of guide RNA designed to bind to a specific genetic sequence roughly 20 letters long.
This guiding process is not always totally accurate, though, and sometimes the Cas9 protein cuts the wrong section, leading to so-called “off target” edits. Depending on what genes are affected, this could potentially have serious side effects for a patient treated with a CRISPR therapy.
So to understand why the system sometimes edits the wrong target, the researchers used a cutting-edge imaging technique called cryo-electron microscopy to take snapshots of the Cas9 protein as it locked onto mismatched sequences.
They found that when the guide RNA binds to the wrong sequence, it normally fails to create the characteristic kink that enables the Cas9 to slice the DNA. But they discovered that when the sequence is correct apart from letters at the 18th and 20th positions, a finger-like structure on the Cas9 protein stabilizes the sequence, correcting its shape and allowing it to be cut.
This was the first time this stabilization process had been observed, and based on this insight the team redesigned the Cas9 protein so that the finger-like structure no longer connects to the mismatched sequence. The resulting protein, which the team named SuperFi-Cas9, was found to be 4,000 times less likely to cut off-target sites than the original Cas9.
This isn’t the first attempt to redesign Cas9 to avoid unwanted edits, but previous approaches have led to significant reductions in the speed of editing. “They’re safer than the naturally occurring Cas9, but it comes at a big cost: They’re going extremely slowly,” said co-first author Jack Bravo.
SuperFi-Cas9, on the other hand, is just as fast as the original protein. While the researchers have so far only tested it out on DNA in test tubes, they’ve already started collaborating with other researchers to try it out in living cells, and the university is looking for industry partners to help commercialize the technology.
While it will likely be some time before such a new technology works its way into the clinic, the research is an important step to creating safer CRISPR therapies that could treat a wide range of diseases in humans.
Image Credit: Jack Bravo/University of Texas at Austin
* This article was originally published at Singularity Hub

0 Comments