A new AI tool opens the door for designer protein drugs that tackle pain, cancer, and brain diseases.
Designing drugs is a bit like playing with Polly Pocket. The vintage toy is a plastic clam shell that contains a multi-bedroom house, a skating rink, a disco dance floor, and other fun scenarios. Kids snap tiny dolls into designated spots so they can spin them around or move them up and down on an elevator. To work, the fit between the doll and its spot has to be perfectly aligned.
Proteins and the drugs targeting them are like this. Each protein has an intricate and unique shape, with areas that grab other molecules to trigger physiological effects. Many of our most powerful drugs—from antibiotics to anti-cancer immunotherapies—are carefully engineered to snap onto proteins and alter their functions. Designing them takes months or years.
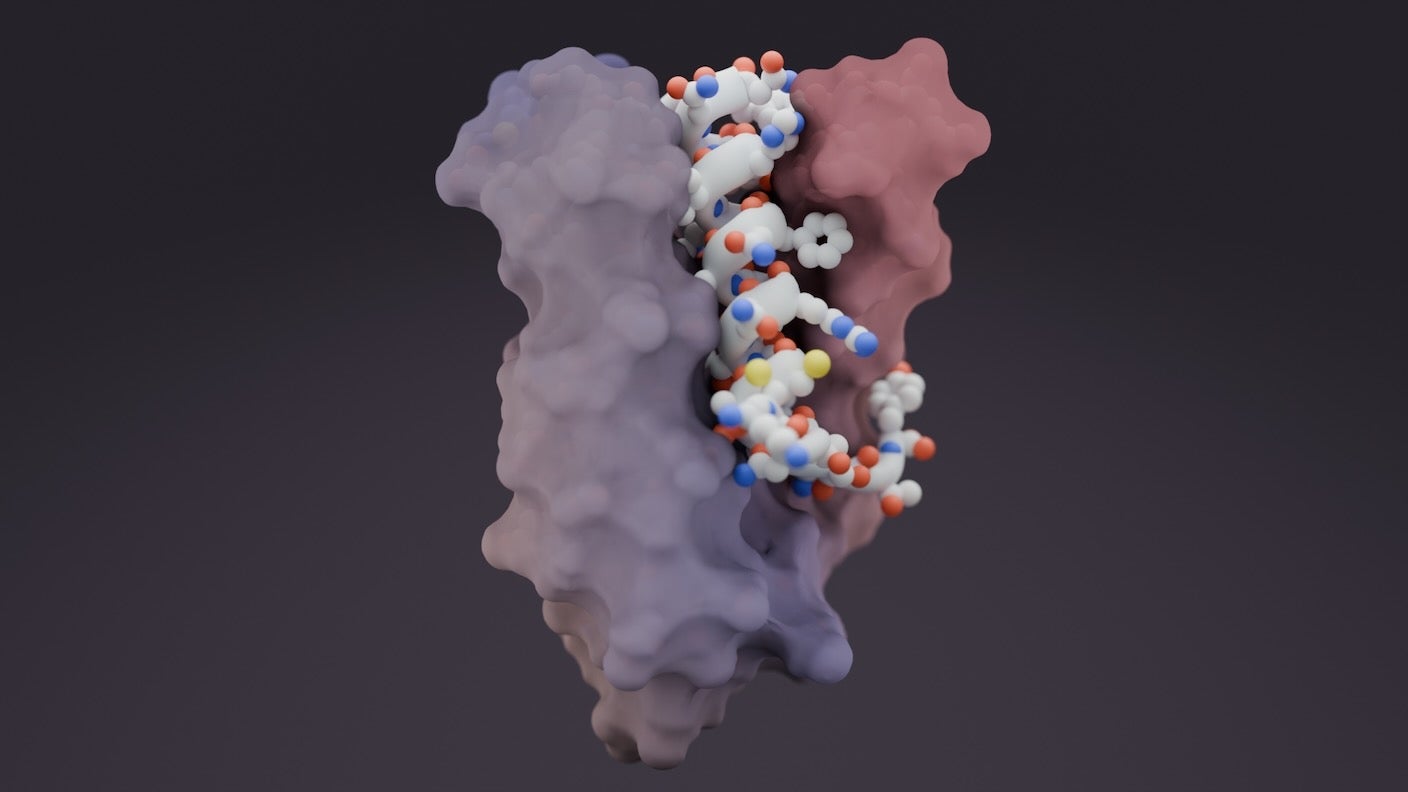
Thanks to AI, it’s now easier to map protein structure, find the hotspots, and design molecules—called “binders”—that grab onto each specific protein pocket.
Here’s where the comparison breaks down. Biological molecules aren’t made of rigid plastic. At least a third of proteins in our bodies contain shape-shifting parts called “intrinsically disordered regions.” Instead of folding into stable 3D structures with pockets for molecules to dock onto, these regions constantly change shape, making it nearly impossible to design binders.
Such proteins are implicated in a variety of diseases, including cancer and Alzheimer’s. Learning to target these tricky shapeshifters could spur a new class of drugs.
This week, a team from the University of Washington led by David Baker introduced a new AI tool that can design binders to grab onto shifty proteins. The AI generated binders to lock onto many previously “undruggable” proteins, including some implicated in cancer.
“Almost half of the human proteome is intrinsically disordered, yet we’ve had no reliable way to drug it. These studies change that by giving scientists everywhere new tools for binding the unstructured half of biology,” said Baker.
A Molecular Dance
Proteins are the workhorses of our bodies. They’re made of chains of molecules called amino acids that fold into complex shapes, like flat or twirly ribbons.
These 3D structures determine interactions with other proteins or drugs. With AI, it’s now possible to predict protein structure and engineer new proteins from scratch. These technologies, though powerful, are mostly limited to stable proteins—those that act a little like Lego blocks—or semi-dynamic proteins that shift from one stable structure to another.
Intrinsically disordered proteins are a different beast. These proteins don’t stabilize, behaving more like jellyfish than Lego blocks. Others contain disordered regions that interact with other proteins to transmit information.
The human proteome—the complete set of proteins in our body—encompasses millions of these interactions that “are responsible for dynamic functions,” wrote Alan Moses and Julie Forman-Kay at the University of Toronto, who were not involved in the study.
Scientists have long eyed these dynamic regions and proteins as targets for drugs. Engineering “jamming” peptides could potentially sever dangerous signals that lead to cancer, senescent “zombie cells,” and a wide range of diseases.
Most AI strategies have focused on proteins with relatively stable pockets for docking. But “because intrinsically disordered regions lack folded binding pockets, it is generally impossible to use existing structure-based machine learning design methods for disordered targets,” wrote Moses and Forman-Kay. Even generative AI that can design binders has struggled here.
Double Team
The new study combined multiple existing approaches into an AI that recognizes disordered proteins and generates binders.
The team first matched repeated structures on the binder and target—a bit like interlocking fingers—to learn about the target’s overall shape. They then shuffled the binder’s features—for example, recombining binding pockets in different configurations—to make a library of binder templates. And finally, they improved on these with an AI technique called diffusion.
In all, the team generated roughly a thousand pockets that “allow for trillions of combinations” that can grab onto wiggly proteins, study author Kejia Wu said in a press release.
As proof of concept, the team built binders for 39 highly diverse disordered proteins. One target, neuropeptide dynorphin A, is crucial for sensing pain. The protein is a popular research subject in pain management, but scientists have struggled to design drugs for it because of its wobbly nature.
The AI-generated binder effectively locked onto dynorphin A’s disordered bits. The protein usually links up with other molecules that either boost or lower its function. Surprisingly, the AI-designed binders stuck to the target better than dynorphin A’s usual protein clique and blocked pain signaling in lab-grown human cells.
New Class of Medicine
Many proteins involved in cancer and brain diseases have disordered regions that are undruggable. Some studies have found small molecules that could target such regions to treat advanced prostate cancer, but successes are few and far between.
As more of these proteins are associated with diseases, binders that change their activity “could have great therapeutic potential,” wrote Moses and Forman-Kay.
For example, new binders could tweak the activity of mysterious droplets called biomolecular condensates floating inside cells. These floating blobs regulate gene expression and immune activation and keep cells healthy when deprived of oxygen and during other stressful moments. Tinkering with them using custom-designed binders could open new ways to influence cellular health for research and clinical use. The binders could also be engineered into antibody-like drugs that compete with pathogens or proteins to stop infections or disease.
They’ll have to be further tested for safety and longevity. But in the future, they could tackle previously undruggable proteins and widen the therapeutic horizon. And they might be used in synthetic biology too. Scientists could design synthetic disordered proteins and custom binders to explore how they work in cells. “This can facilitate a wide range of experimental and translational applications that were not previously accessible,” wrote Moses and Forman-Kay.
The post AI-Designed Drugs Can Now Target Previously ‘Undruggable’ Proteins in Cancer and Alzheimer’s appeared first on SingularityHub.
* This article was originally published at Singularity Hub

0 Comments